WormBase ID: WBGene00015497 WormBase description: Is predicted to have DNA-binding transcription factor activity and RNA polymerase II regulatory region sequence-specific DNA binding activity. Is involved in positive regulation of fatty acid beta-oxidation by octopamine signaling pathway; positive regulation of fatty acid beta-oxidation by serotonin receptor signaling pathway; and positive regulation of transcription by RNA polymerase II. Is expressed in several structures, including body wall musculature; excretory gland cell; intestine; pharynx; and vulval muscle. |
Reporter verification: Whole-genome sequencing indicated that the GFP tags another TF, NHR-76 rather than F22D6.2 |
Strain name: SYS558 Cross: OP399 x JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; wgIs399 (nhr-76::TY1::EGFP::3xFLAG + unc-119(+)) |
Expression pattern: Expressed after bean stage |
Lineage expression pattern 350 stage 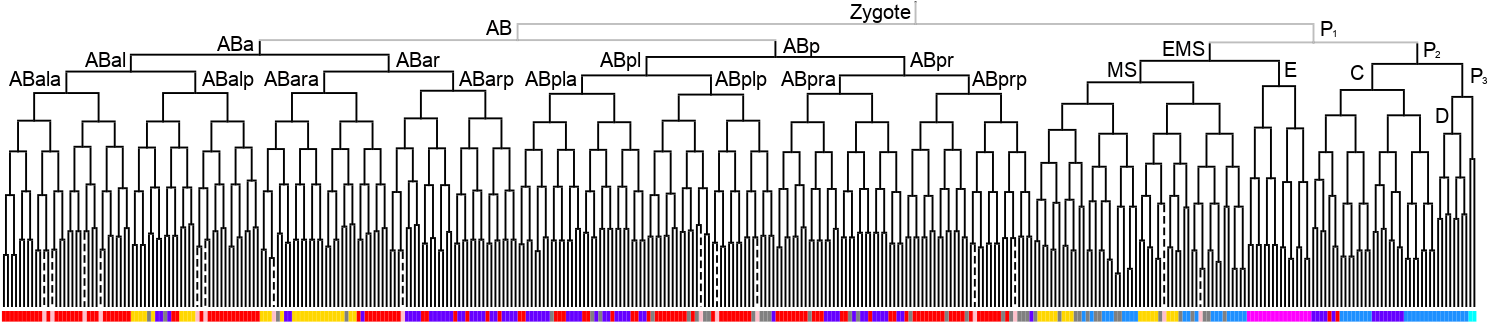 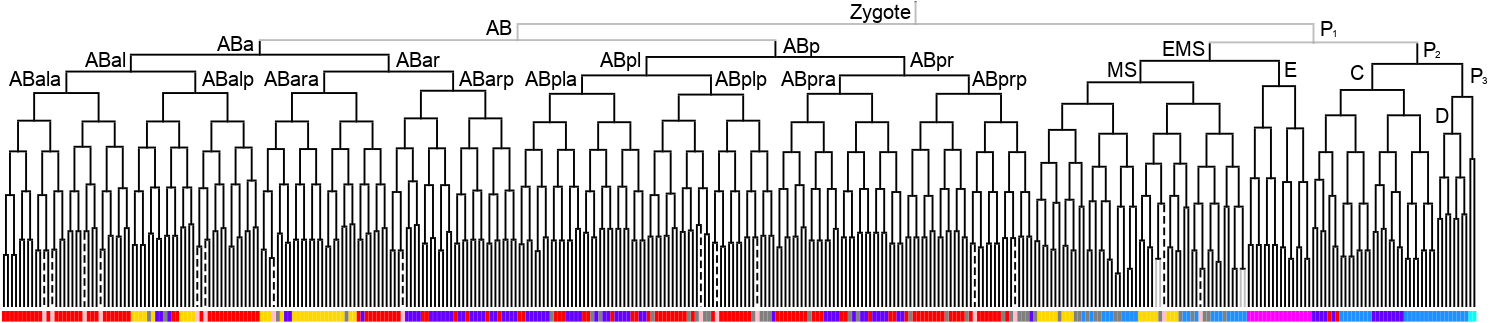 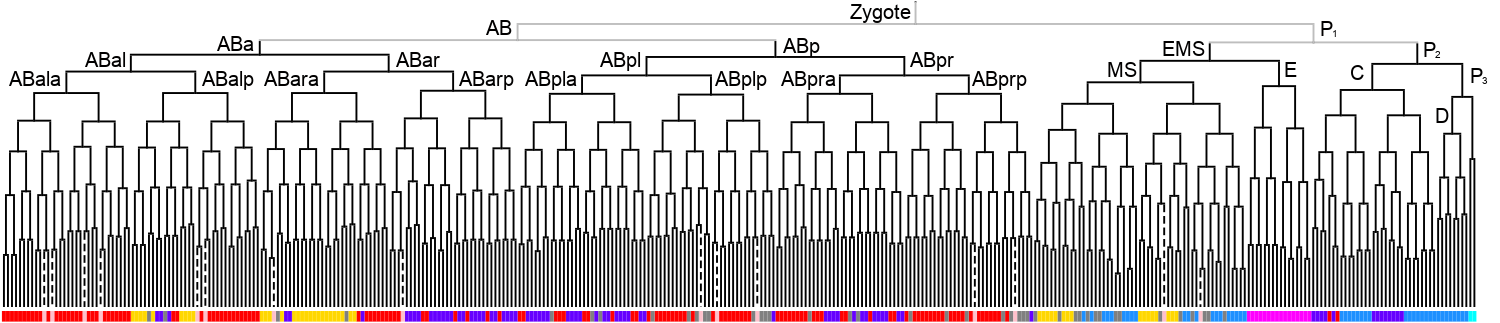 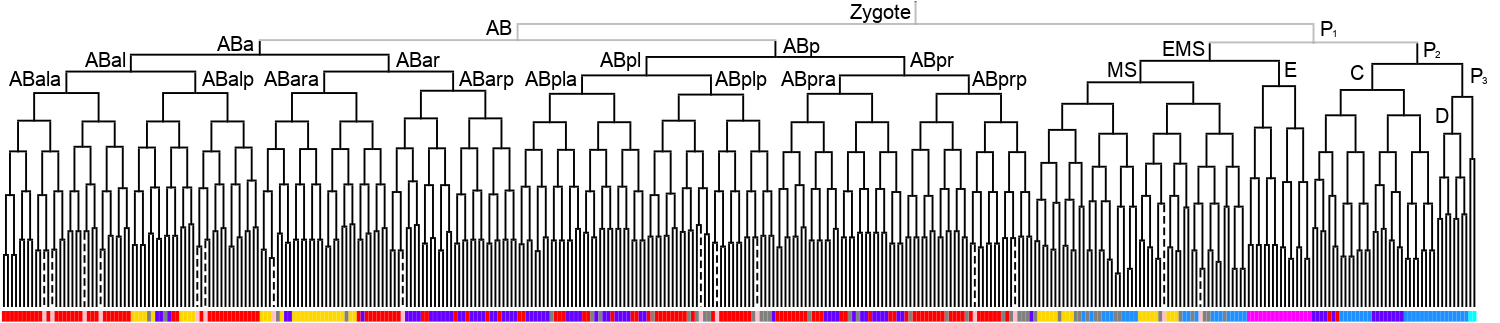 |
600 stage 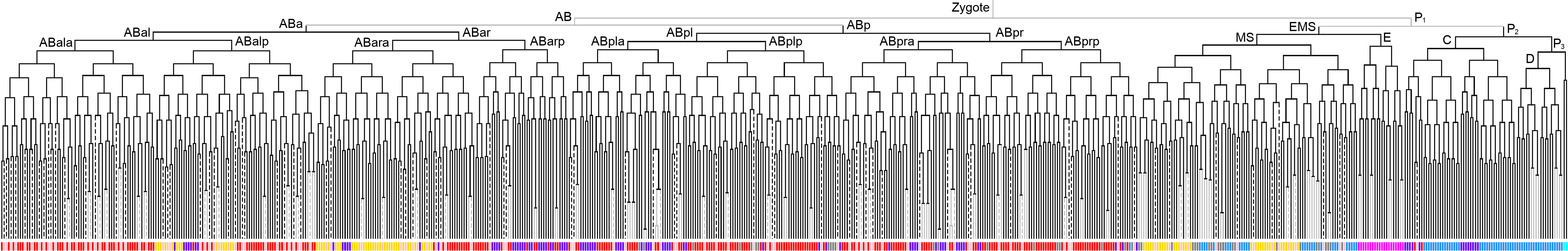 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): NA Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) |
|
Targeted by: CEH-31 |