WormBase ID: WBGene00004334
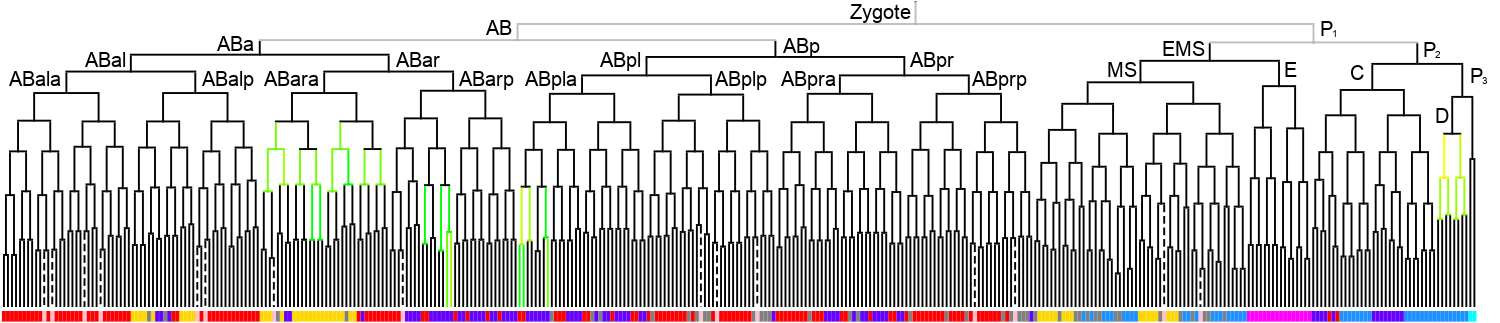
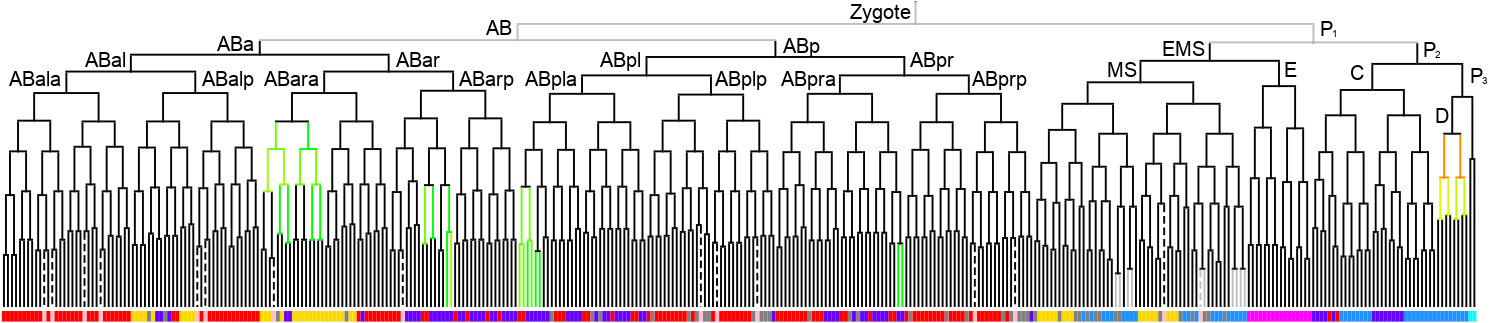
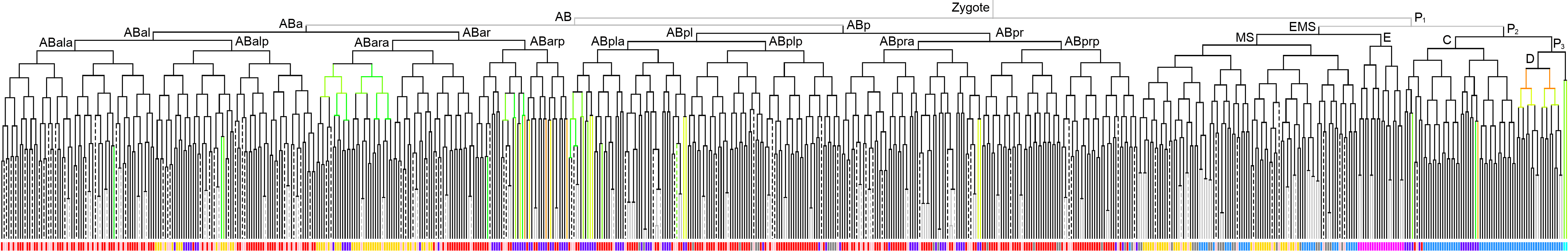
WormBase description: Is predicted to have protein dimerization activity. Is involved in several processes, including epidermal cell fate specification; negative regulation of transcription by RNA polymerase II; and nematode male tail tip morphogenesis. Localizes to nucleus. Is expressed in several structures, including E lineage cell; embryonic cell; intestine; mc3V; and pharyngeal cell.
|
|
KI information: mNeonGreen knockin at N-terminus
sgRNA1: GTACTGATGAGGACCATTTC
sgRNA2: TCATAACAGAAAAACATCAC
5'HR: GCATGCATCTTCTTCTCATTTTTTTGTTTGTTTTTTAGTTCCAGTATTTACTTTAGCTGTTTTGAGATTTTGAGAAGTGTATTTAGTATACATTCCTAATTATTTTTTGCCCTAAATTCTTTAGAGTCTTACAGTAGCAAATCTACAGTAGTCTATAGCTATGGTTAAGAGTTTCAGCTACAGTAGCAACATAGTTCTACCTCACCAGAAATAGTATTGCTACTGTAGCGCTTCCAATTTCTAGTTTTCCTTAAATTAACCAATTCTCACTTCATTTTAACATTCTTCCCCCGAATTCTCTATGTCTCACCTCACTATCAGTCTTCCAAATTTTCACCTCAAGTACAGTACATCTGAAACTCTAACACCTCCCCATGCATTTTACTGCCTGTAGATCCCTCTTGTTTACCAACTTCTTTCATGTTCTTCCGGTCTGCCGTTTCACTTCCTTTTCTCACCTTGTCGGACCCCTAGCTCTCCATTAAAGGATTAGTACCGCCTGAGCATCCGCCCGGCTCTTCACAGTGTCGCCTCTCACCAGGGGTTATCAAACCAATATGGTCAAGAGTTGTGGGAAACTGTGTCATGACTCAAGTGCTTTTTTCCCATGCACACTTATCAAATGATTCCCACACTGATAACTCGGCACGTCTTTTGTGGTTGATAGTGATGGGAACCATTGGGATATAATTTTTTTTGGCGAAACGACACAAAAAGTGGGTTTTTAGGGTTTAAGGCAATTAAATTTCTACGAATAAGATTCTAATCTAGAAAAAGAAGGCGTGCTGTAGAAGTGAGGGGTACTGTAAATCGCCACGCTCCCTCTTCCGAATGGTTTATGACTTTTCTCTATATCTCCTTCTCTTTCTCTATATTTCTCAAATTCCGCAACATTTCTCCCTTCTTGATTTTGTTCATTTACAAATCGAGCTATAAAAATCAATAAAAACTTAATTTTTTTTCCAGAAGGTACC
3'HR: CTGCAGATGGTCCTCATCAGTACCCCACCACCAGCTTACGCTCATAACAGAAAAACTTCACAAGAGAAGAAACGACGAGATGAGATTAATGCAAAGATCAAGGAGCTTCAACTATTGATTCAAAATGAATCGGATAATGAGAAAATGACGTGAGTTTTCTATTCGTTTAGTGTTATCTTCATTCAAATCTTACTAATTTTCAGCCAAGGTGATGTCTTAAACCGTGCAGTCGAAGTGGTCAGTCGCATGGAAACCGAATCACCTGGACCATCATCAAATCCAAATCGCAAAGGATTCTTTGACGGATTCCGATCAATCGAGTCGCTCACTTATTCCTTCATCAAATCCCTGGGCGTCAATTCAGATGTCTGTCAGGATTTTGTGCAACGTGCCAAGCAGTTTTTCGATCGAGAACGTTCATCTTTGCTGAGCACTGTATCTGGAAAATCAAAGAGAAGATCTGAGTCGGAAATTCTCCACTCTTCAATGAGCTATAGATCCCAGTCAAGCTCTCCATCGACATCGGAATCTGGAATCACAATTGATAGAAAGGAAGTGAAAAAGAATAGAGAGCAAGATAGAAGAGATCGTCAGGGAGAAGCTTTTGATGCTTTGAAGAACTTTATCATTGAGAACAAGCTCATGACATCTCATCAAGTGTAAGCTTAAATTTTCCCAATACCATAAATTAAACTTTTCCTTTTTTCAGCGAAAAAATGCAACGCTTGAACACCCTGGATATCATAATTGCCTACATTCAAAACAAGAAGCACAATTTTGTTAGTCGAAGCGACCAGGAGCAGTCACTCTATGCACATGCAATTGCAGAGGGAAAGAAGACTGCAAAAAACATTGCCTTCCAATTTTTCAAGGTGAGTTGTCAGGAACTTCGAGAAGATTTTTTTAAATGTTATCAATATATTTTCAGAGCGACCGTCACCTCGTTGTCCGATGCGCAGACCTTACTAGT
KI verification primers: REF-1-NKIF: 5'-ATTAGTCAACAAGTGGGGGCTATC-3' REF-1-NKIR: 5'-AGTGAATATGATGGAGACTGTTGAGAT-3' Download
|
Inferred TF relationships
Based on expression similarity (strong, medium, weak): LSL-1, RNT-1, CEH-16, HND-1, NOB-1, NHR-12, NHR-23, GMEB-1, LIN-26, ELT-1, HAM-2, NHR-25, CEH-39, HBL-1, TBX-11, NFI-1, CEH-74
Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined)
Targeting: no-data
Targeted by: ELT-1, CEH-83, ELT-2, HLH-1, TBX-8, UNC-55, DSC-1 |